We presented echinopscis, an overview and our design decisions in the symposium “The evolving landscape of biodiversity informatics: bringing actionable practices and tools to practitioners” in TaxonWorks Together 2023
About Taxonworks
TaxonWorks (taxonworks.org) is a biodiversity informatics software platform created by a team at the University of Illinois, described as:
an integrated web-based workbench for taxonomists and biodiversity scientists. It allows you to capture, organize, and enrich your data; share it with collaborators; and package it for analysis and publication
“TaxonWorks Together” (together.taxonworks.org)is an annual online event which showcases the platform and is designed to help users share their use cases and experience, and influence future direction.
Echinopscis presentation
Overview
We presented echinopscis in the symposium session “The evolving landscape of biodiversity informatics: bringing actionable practices and tools to practitioners”. As well as a general overview introduction to echinopscis, this talk emphasised the design principles in the project:
- Data are stored locally - low barrier to initial use and experimentation
- Focus on reusable skills - any skills development (eg markdown authoring, github proficiency) required to become more effective with echinopscis should be an investment that can also be used elsewhere in other projects and platforms - not a single app “time sink”.
Relations to TaxonWorks
Local storage means that there is an opportunity to use (i) as a starting point - users could initially work in echinopscis and then “upscale” into taxonworks and (ii) as a “safe haven” - to show that a data download from taxonworks could be used in a standalone echinopscis environment.
TaxonWorks also represents a valuable source of data that echinopscis should be able to interact with.
More fundamentally - the use of technologies like github pages (used for TaxonPages), and skills like markdown authoring etc are shared between the two projects and therefore learning resources could be developed collaboratively.
Discussion points
We then posed a couple of discussion points - (i) widescale adoption of the reconciliation API and (ii) enabling the navigation of linked data.
Reconciliation API (or “strings to things”): the echinopscis demo shows multiple workflows where a user translates a piece of text (like a specimen reference) into an entity managed by an external authority (like a specimen record). Many users may be aware of the reconciliation process in Open Refine (a tool for manipulating messy data). This Open Refine Reconciliation API is now documented as a standard and can be implemented more widely. Some current data provider examples include:
- Bionomia: bionomia.net/reconcile
- Catalogue of Life: github.com/CatalogueOfLife/backend/issues/1265
- Datasette (generic): github.com/drkane/datasette-reconcile And some examples of data consumers (developers of taxonomic / data manipulation tools) include:
- Open Refine: openrefine.org/docs/manual/reconciling
- Python: github.com/jvfe/reconciler
So - could we build an Obsidian plugin to faciliate turning “strings to things”?
Navigating linked data “entity explosion” is a browser plugin that shows links from Wikidata based on the currently loaded page. Here is an example of an author profile page from the International Plant Names Index, showing data retrieved from WikiData via this extension:
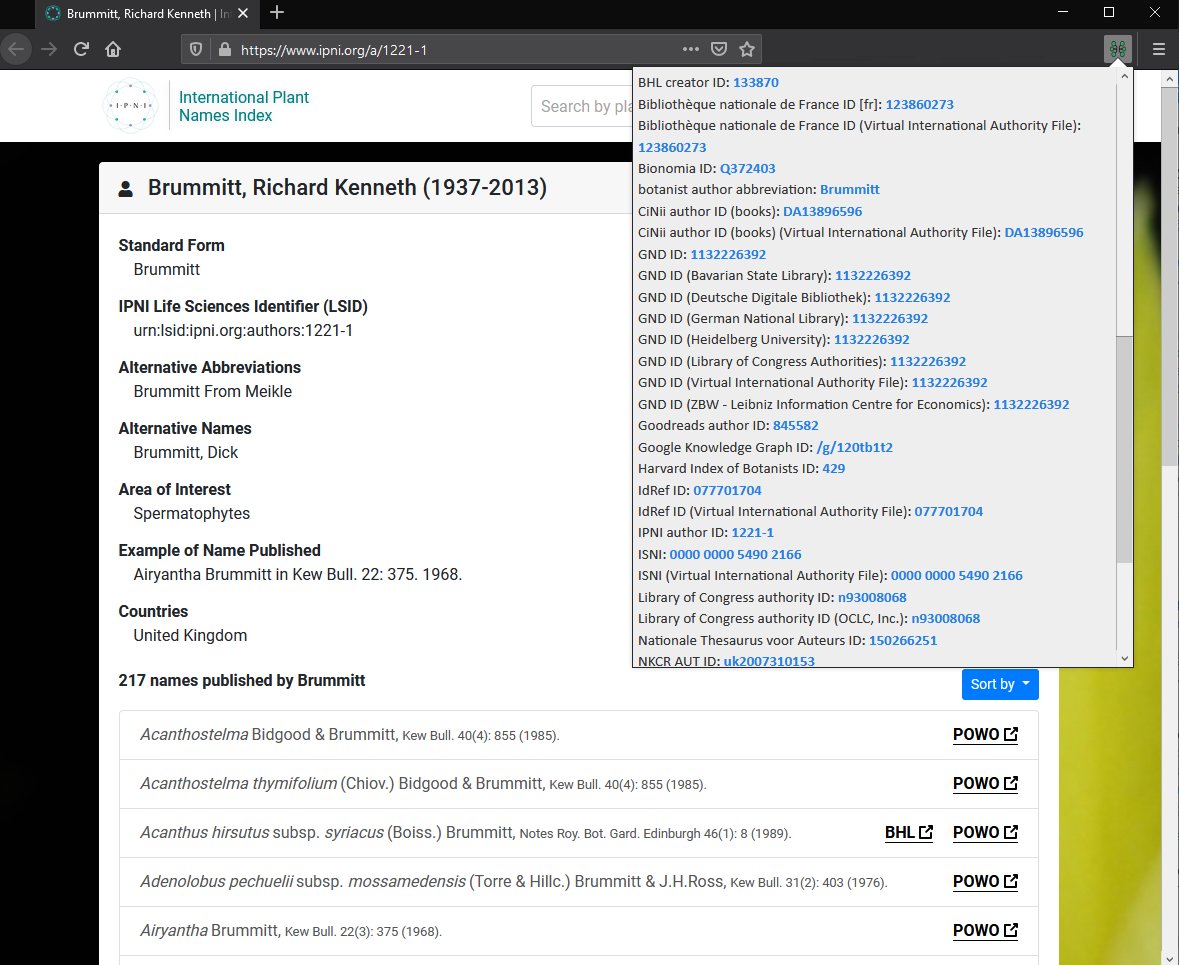
Could we develop equivalents for:
- Obsidian - using an ID stored in page frontmatter to run a Wikidata query and show useful links in a sidebar
- Datasette
- or a “panel” for use in TaxonPages or in the GBIF hosted portals environment
Useful links
- Slidedeck, talk recording (link TBC)
- echinopscis discussion board: github.com/orgs/echinopscis/discussions