Video and annotated slide deck for lightning talk “Open science tools: Supporting hands-on creation of the "digital extended specimen"” from the Biodiversity Information Standards 2022 conference in Sofia, Bulgaria. Abstract: doi:10.3897/biss.6.91123

Work is a collaboration between Nicky Nicolson (NN) and Eve Lucas (EL) both from Kew Science

Personal context: NN transitioned from software development into research, and is interested in how software development practices can be used in research. Institutionally, we have a commitment to accelerate taxonomy using digital practices, and we started a collaborative project to explore this, using institutional knowledge from the e-taxonomic approaches, but exploring what technical advances could be used in e-taxonomy today.
Jump to 0m31s
Moving up to the community in which we work - one of our aims in biodiversity informatics is to build a digital extended specimen, integrating specimens and their derived data across multiple infrastructures, allowing the investigate of wider research questions. A healthy community will support a range of different approaches as we determine how to reach this aim. We can envisage these appraoches as a spectrum from large scale computation approaches that operate on large volumes of aggregated data (like the GBIF clustering work), to a distributed set of lightweight tools that allow users to conduct link construction in context, closer to their day to day work.
Jump to 1m33s
We’re focussing on tools for researchers in this project, though we aim that data generated here will feed into more automated approaches - both by providing expert generated links that can be used as training data, and by allowing experts to verify the outputs of machine learning processes. We also want to try to help make discussions about the digital extended specimen a little more concrete, by having actual examples and workflows that can be used as the basis for discussions and planning.
Jump to 2m40s
We’re basing this work on an existing piece of software - Obsidian. We said that we’re trying to bring across useful working practices from software development to research management - reuse is a core principle in software development and here we’re trying to build on an existing toolset and supportive community to allow us to make faster progress. We have a biodiversity crisis: faster progress - and wider participation - is vital.
Jump to 2m54s
These attributes should be familiar as they are shared with a tool which we have adopted with some success in our community: Open Refine. We’ve seen that the use of a generic tool, with focussed technical contributions and teaching resources has allowed us to democratize data cleaning. We’d like to investigate if we can build on Obsidian and democratize data linkage.
Jump to 5m53s
What we’ve done - extended Obsidian for specimen research, by developing a set of plugins that allow a researcher to access data: specimens from GBIF, names from the International Plant Names Index, collections from the Global Registry of Scientific Collections, people from Bionomia and literature from crossref. The demo software includes a worked example showing the creation of links, and spatial and network exploration of linked data. The demo also shows how key entities (specimens) can be cited in new work, promoting open science working practices.
Jump to 7m10s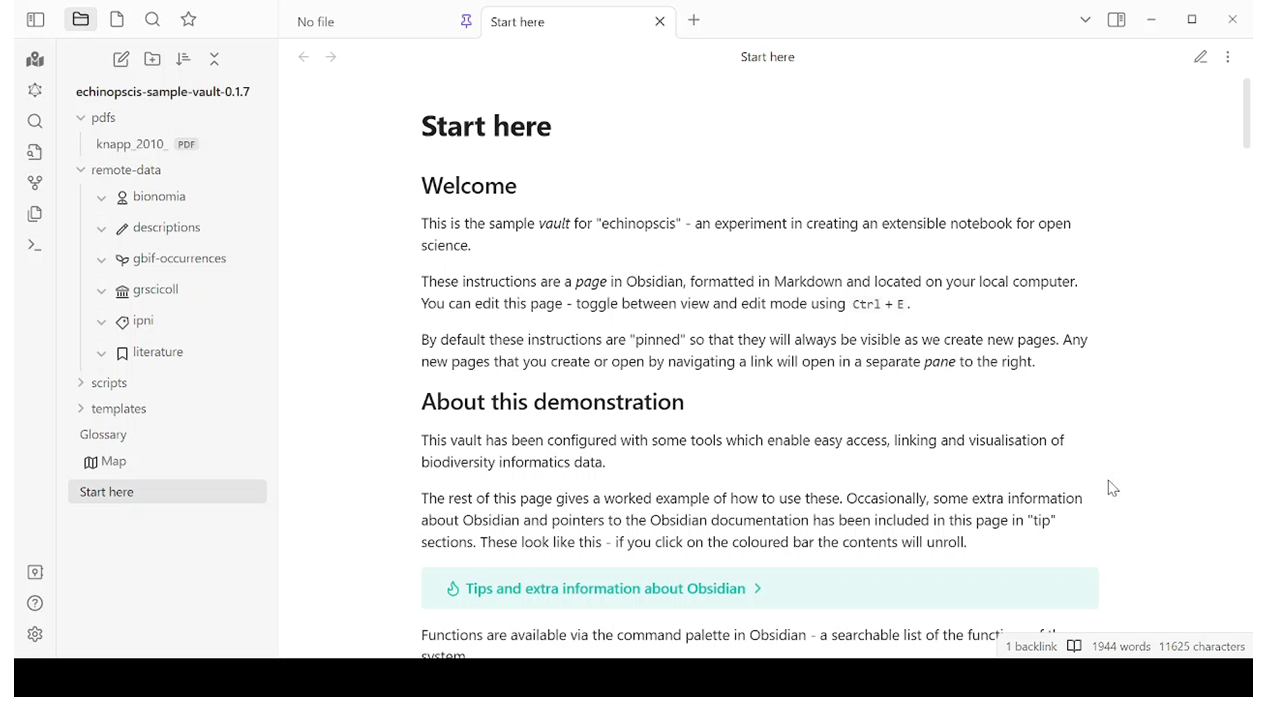
The demo includes an overview of how we can work with text data, highlighting a scientific name and searching for it in IPNI, retrieving name publication data including type citation data, search for type specimens in GBIF, search for collector profiles in Bionomia, using the DOI attached to the IPNI record to get bibliographic information back from crossref. The demo shows how the Obsidian user interface enables a user to link up data and to explore data visually, using a network of links
Jump to 8m11s
We’ve proposed a 4 phase outline roadmap - work demonstrated so far fits into phase 1 - a personal research environment. We think that we can support more advanced use cases in phases 2-4, such as the production of research websites using static site generators (here we see conceptual similarity with the GBIF hosted portal programme), production of documents with embedded specimen references, and production of datasets for harvesting by data aggregators. We’re participating in Open Life Sciences and we will revise our roadmap as we build a community around this project.
Jump to 12m10s
Some links to more information and contact details.
- Echinopscis:
- Project site (background, installation instructions, roadmap, ideas for contributions): https://echinopscis.github.io
- Code repositories and documentation: https://github.com/echinopscis
- Obsidian:
You can find me on github, my handle is @nickynicolson. Since I wrote these slides I’ve moved from twitter (where some old stuff is still available at @nickynicolson) to Mastodon where I am @nickynicolson@mastodon.social
Jump to 14m41s